Feature
Published May 31, 2013

Molecules are arguably the most fidgety things in the universe. Their atoms are in constant motion, making slight position adjustments in timescales that start in femtoseconds—or one quadrillionth of a second.
These short-scale atomic transitions are the starting point of the varying molecular conformations that drive vital biological movements, like the opening and closing of protein channels that trigger a heart beat, or the movement of a molecule through a cellular membrane.
[[It’s a common technique for computational laboratories to map the steps of molecular movements by piecing together billions of femtosecond long atomic place changes into a microsecond long snapshot. But for many important biological processes, microseconds are simply not long enough.
DNA polymerases, the enzymes that piece together DNA take milliseconds—thousands of times longer than microseconds—to add a new nucleotide, and the diffusion of some substances through cell membranes can take hours. A few microsecond snapshots hardly give a clear picture of processes like these.
What they need is a timekeeper that can keep up for the long haul. Read more.]]
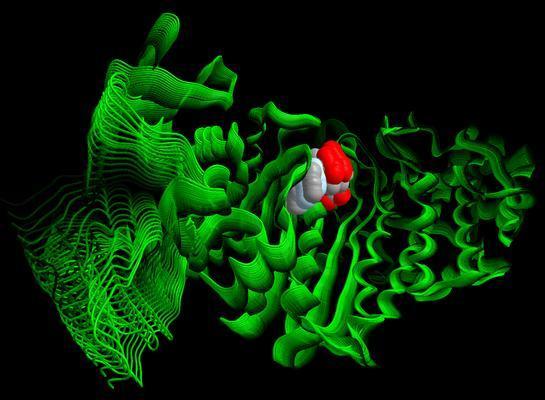 Milestoning, an algorithm developed by ICES’ Ron Elber, is the ultimate timekeeper. By breaking the movements of molecules into discrete patches of shorter time that can be pieced back together, Elber creates computer simulations that investigate molecular movement in the time scale that they actually occur, whether that happens to be milliseconds or hours.
Milestoning, an algorithm developed by ICES’ Ron Elber, is the ultimate timekeeper. By breaking the movements of molecules into discrete patches of shorter time that can be pieced back together, Elber creates computer simulations that investigate molecular movement in the time scale that they actually occur, whether that happens to be milliseconds or hours.
“Milestoning allows us to match timescales that are important to biology, and which other computational methods have not been able to do,” said Elber, director of the ICES Center for Computational Life Sciences and Biology. “This theory has opened the way to studying many, many other processes.”
One of these processes is the movement of myosin, the “molecular machine” responsible for muscle contraction and relaxation. An action as simple as raising a hand takes trillions of myosin proteins working in tandem.
Using Milestoning, Elber developed a simulation that accounts for the movement of every atom in the myosin protein and how it moves during the millisecond-long movement of the protein as it contracts. Fittingly, the overall movement resembles a flexing bicep.
“This is a collection of many, many small steps leading to a slower process,” said Elber, referring to how the fleeting atomic movements ultimately influence the movement the entire myosin protein.
The reason that conventional methods can’t get passed the millisecond time scale required for simulations like the myosin cycle is a fundamental difference in the arrangement of the steps that make up a molecular movement and the arrangement of processors used in modern high-performance computers. The steps are sequential, while the processors work in parallel—computing multiple parts of data input at a single time.
And while the calculations required for a single femtosecond step can be organized to be computed on parallel systems, the computing resources, and time required to do so are enormous, hence the conventional limitation to the microsecond time scale.
Elber is able to get around these limitations by turning the steps from a solid timeline of events into a “connect-the-dots” puzzle of sorts by a applying a statistical interpolation.
This method uses a selection of atomic confirmations at representative times—these are the “milestones” that give the method its namesake—and applies statistical and mechanic theory to estimate the atomic trajectories that take place between the spatial gaps that separate one point from another. The number of milestones varies from several tens to tens of thousands, depending on the process.
“It is like reading a really thick mystery novel. You can read it one page at a time or you can “cheat” and read a few pages in the middle and at the end and then interpolate what happened in the middle,” said Elber. It took 200 milestones to create the simulation showing myosin’s contraction/relaxation cycle.
Breaking steps into segments separated by milestones aids in the computing process, too. The milestones and the gaps between them are easily broken up for parallel computing, and later pieced back together—similar to assigning different chapters of a book to read to a group of people, and then conferencing about the entire story at a later date.
Milestones are assigned to the parts of the molecular process that have the most action, like when a substrate binds or there’s a significant shape change, says Elber, which helps increase the accuracy of the inputs that inform the theory that fills in the gaps.
Elber has used Milestoning to investigate molecular processes that work on time scales that are orders of magnitude apart, from the hours long diffusion of molecules through a cell membrane, to the microsecond long activation of enzymes by their substrate.
One research endeavor examined cellular transport in real time to understand more about cellular life in ancient times.
The membranes of most modern day cells are embedded with channels and pumps to assist the transport of materials in and out of the cell. But when cellular life was just getting its start on Earth, it’s likely all the cells had was a sealed membrane and molecules had to pass directly though it.
Using Milestoning, Elber timed how long it took for different essential molecules to pass through a membrane. And while sugar was able to pass through in microseconds, amino acids—the building blocks of proteins—took hours to diffuse, too long to sustain protein production essential for cell life. The sluggish pace of amino acid diffusion suggests that some other method must have existed to get the amino acids into the cell more efficiently.
“We were given an indication of possible processes that can happen during the early steps of evolution,” said Elber. The research techniques can potentially be applied outside of the primordial soup, too, for the more modern need of understanding the uptake of various medications by cells.
 In another research project Elber teamed up with Ken Johnson, a UT biochemistry professor, to analyze how the molecular movements of HIV’s reverse transcriptase enzyme influence which nucleotides are incorporated into a DNA strand. Reverse transcriptase is vital for the survival of HIV because it uses the virus native RNA genome as a template to create DNA copies, which are integrated into the human genome for virus replication.
In another research project Elber teamed up with Ken Johnson, a UT biochemistry professor, to analyze how the molecular movements of HIV’s reverse transcriptase enzyme influence which nucleotides are incorporated into a DNA strand. Reverse transcriptase is vital for the survival of HIV because it uses the virus native RNA genome as a template to create DNA copies, which are integrated into the human genome for virus replication.
The enzyme movements modeled by Elber gave reaction rates very similar to those produced experimentally by Johnson in the lab, lending support that Elber’s simulation was correct. Knowing more about how the molecular conformation along each step of the enzyme’s process can potentially help other researchers in developing methods to interrupt the process, said Elber.
“In principle, you can perturb the protein so it doesn’t work at different stages. We’re just not sure which stage will be the easiest to interrupt.”
At the moment, Elber has returned his research focus in part to myosin. Now that he knows in detail how the molecule moves he’s using Milestoning to test what it takes to break it.
“There’s a question about the mechanical strength. How much can myosin suffer and not crack?,” said Elber. “ In the same way people test elements of a machine, or a car, we are now testing this piece of a mechanical system.”
In the body, a single myosin protein handles about 10 piconewtons of force. But Elber’s simulation revealed that maximum force a single myosin protein could bear before its bonds broke beyond repair was 20 piconewtons. And when force was initially applied, the molecule did not waver under the extra load. Unexpectedly, said Elber, it became “stronger,” or in molecular terms, entered into a more stable conformation.
“It’s more stable when you pull on it a little bit,” said Elber. “It’s only when you pull really hard that it breaks.”
The reason for the increased strength, says Elber, is the extra force causes the molecule to become more rigid, locking the myosin into place and enabling it to better handle the extra force.
Elber started developing Milestoning a decade ago while at Cornell University. Now, at ICES, his technique works along the many algorithms employed, and being developed by, ICES scientists to investigate biological phenomena and structure. Elber collaborates, and checks findings with experimental scientists. But research in biology is more and more dependent on computational methods to investigate worlds that are difficult to access any other way.
“We are developing a very general tool for the community to use on biochemical and biophysical molecular events with a long time scale,” said Elber. “More people are using our ideas and technology to address a wide range of problems.”
Article by Monica Kortsha