Feature
Published March 5, 2013
[[African sleeping sickness is a disease that starts out in a small world. It is a condition transmitted by the bite of the tiny tsetse fly and caused by a microscopic parasite that goes from living in the fly’s gut to the human bloodstream, where it can wreak havoc on the body. ICES’s Chandrajit Bajaj, a professor in computational visualization, has helped provide a view of yet an even smaller structure vital to the life cycle of the parasite: the ribosome. Read more.]]
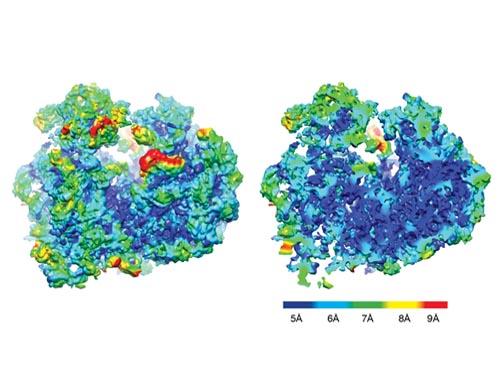
Ribosomes are essential organelles for every living creature because they translate the genetic code into proteins a cell uses to perform a host of vital functions. Bajaj, who directs the Computational Visualization Center at ICES helped visualize the first high-resolution and 3D model of the Trypanosoma brucei ribosome using computational methods.
The research, conducted jointly with structural biologists and biophysicists from Columbia University, the New York State Department of Health and the Université de Strasbourg in France, was published in Nature online in February.
Knowing the structure is the first step to inspiring all sorts of further avenues of research, including more effective medical treatments for the disease.
“First we try to reconstruct the structure, obtain all the atomistic constituents of the makeup. Once that is vetted properly we embark on the next questions of elucidating structure/function relationships and potential cures for the disease,” said Bajaj, who also holds the Computational Applied Mathematics Chair in Visualization.
The organism that causes sleeping sickness belongs to a larger group of protozoa called kinetoplastids. They’re a varied bunch, including free-living as well as plenty of parasitic varieties. What they do have in common is a diverse and unique ribosomal structure when compared to other eukaryotes, whose ribosomes differ very little from one species to the next.
While all eukaryotic ribosomes have expansion segments, a name given to certain inserts of RNA code, the sleeping sickness ribosome was found to have several segments double the size of any others that have been observed. These segments in turn form four “inter-subunit bridges,” another unique feature, that connect different parts of the ribosome together.
“These expansion segments for some reason are way larger in this parasite than in any other known eukaryotic ribosome,” said Yasar Hashem, a post-doctoral researcher in Joachim Frank’s lab at Columbia University who headed the research collaboration. “And they’re not only larger, but they form very peculiar structures that have not been seen in eukaryotic ribosomes.”
Some of the ribosomal proteins were found to have novel structures, too, including helical extension that stabilized the large expansion segments as they reach and weave throughout the entire ribosome.
Collectively, the features and others are “extreme” in their diversity, said the research article. But being able to accurately map and describe the topography of an organelle on the molecular scale in the first place takes some special tools and software. A knack for puzzles helps, too.
The first step in the high-tech process was a photo shoot. The ribosomes, after having their poses fixed by a -180°C ethane dip, had their photos snapped hundreds of thousands of times under a microscope with near atomic resolution. The result was two-dimensional images of the ribosome at nearly every angle.
To go from a flat photo gallery to the 3D visualization required making a composite, said Bajaj. But it wasn’t as simple as combining all the photos together panorama style.
Reconstructing the ribosome’s three-dimensional shape from the 2D pictures required first relatively orienting the macromolecules in perspective to the microscope lens by applying methods similar to that used by astronomers to map the rotations and orbits of distant planets.
“You recover the relative orientation or the unknown relative pose of these images with methods similar to how one may recover the rotations and the orbits of different planets and stars because they are tilted in various respects toward your telescope,” said Bajaj. “So the microscope is similar to fixing the orientation of the telescope and you’re capturing the relative orientations of millions of nanoscale particles almost all at different orientations.”
Once the 3D picture was constructed from the appropriately aligned 2D images, the individual RNA and protein domains had to be deciphered and disentangled from the larger molecular structure.
Bajaj and his team developed two programs to help make sense of the photos, turning the tangles into recognizable individual molecules and identifiable molecular sub-domains.
The first, VolumeRover (in conjunction with the Frank lab’s software called SPIDER) helped to turn the 2D photos into 3D volumetric images, as well as distinguish the individual proteins and RNA molecules present in the structure. TexMol aided in the 3D puzzle piecing part. It compared the ribosome to known structures, including E.Coli and additional protozoa, to identify what structures were shared and what parts were new. Further, known atomistic structures of the shared individual proteins and RNA molecules were automatically fitted together and into the reconstructed 3D images.
“You use various quantitative metrics to say you have a good alignment and a good fit, and then you go back to see there are some densities that are missing, and that’s how you also discover structural differences, said Bajaj. “It’s like putting together a 3D jigsaw puzzle, when you also know that may not have all the pieces.”
In addition to programs developed at ICES, Bajaj used programs from colleagues at Columbia and other institutions, to cross reference results for maximum accuracy. It’s a necessary step that Bajaj refers to as a tour de force that keeps discoveries from turning out to be errors.
“Whenever you’re discovering structures which are unknown, finding needles in the haystack, you have to be careful that you have the right set of controls and you’re not predicting erroneous needles,” said Bajaj.
The result of the research is a precise and dynamic range of ribosomal models, which are likely to serve and inspire dozens of research streams, and potential medical treatments, to come. And the impacts range beyond the 30,000 people thought to be currently infected with sleeping sickness.
“If you find common features from these parasites you could narrow it down to a few essential features that are always there and target these features,” said Hashem. “This could be used in the process of developing new drugs.” And new drugs are needed, as current ones could prove fatal to the patient.
The small scale, it seems, can make a big difference. Whether it’s the near invisible transmission of a parasite from a fly to human, or enabling potential cures by visualizing even smaller structures yet.
Article by Monica Kortsha